DasR
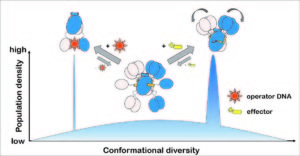
DasR from Streptomyces coelicolor belongs to the GntR/HutC family of bacterial repressors and its DNA-binding properties are modulated by phosphorylated sugars. Although the GntR proteins represent a large family of repressors, only little is known about the allosteric mechanism that enables their function. By solving multiple crystal structures of DasR in different functional states in combination with molecular dynamics (MD) simulations and structural data available from additional family members we were able to show that the allosteric behavior of DasR and possibly many other GntR/HutC family members is best described by a conformational selection model. Effector binding to the effector-binding domains (EBDs) of DasR significantly reorganizes the atomic structure of the latter. However, rather than freezing the orientation of the DNA-binding domains (DBDs), the effector-induced formation of β-strand β* in the DBD-EBD-linker segment merely appears to take the DBDs ‚on a shorter leash‘ thereby impeding the ‚downwards‘ positioning of the DBDs that is necessary for a concerted binding of two DBDs of DasR to operator DNA.
PLoS One. 2016; 11: e0157691 (PMID: 27337024).